Disease, Research
The ‘thrilling’ discovery of the Y4 leukotoxin and the finding of Aggregatibacter (Actinobacillus) actinomycetemcomitans
27 January 2024
The discovery of Y4 leukotoxicity and the subsequent finding of JP2 as a “super” leukotoxin producer were key moments in the development of periodontal science and the understanding of the inflammatory response to bacteria. This account of the discovery of the Y4 isolate leukotoxin leading to the identification of Aggregatibacter (Actinobacillus) actinomycetemcomitans focuses on the work of a research group at the University of Pennsylvania (USA) between 1974 and 1981. Members of the research team – Pierre C. Baehni, William P. McArthur, Norton S. Taichman, and Chi-Cheng Tsai – tell the story.
The discovery of Y4 leukotoxicity and the subsequent finding of JP2 as a “super” leukotoxin producer were truly extraordinary events for our team. In the mid-1970s, we started to explore the interactions between bacteria and polymorphonuclear leukocytes (PMNs) that may be relevant in the pathogenesis of periodontal disease.
In the 1960s, periodontology was an established and long-recognised speciality of dentistry. Gingivitis, periodontitis, and periodontosis (later to be known as juvenile periodontitis and then, under the current classification, localised aggressive periodontitis) were recognised as distinct inflammatory conditions of the periodontium, and it was generally accepted that the accumulation of dental plaque was responsible for these diseases. But there was still a limited understanding of the microbial components of dental plaque and how they caused these inflammatory diseases.
Boosted by funding from the National Institute of Dental Research (NIDR), research centres started to address these critical issues and major advances were made in oral microbiology with the introduction of new cultivation techniques, derived from studies on the intestinal flora. Studies showed that anaerobic microorganisms constitute a large segment of subgingival plaque and suggested that the oral microbial flora might comprise between 200 and 250 different microbial species. It was also apparent that the bacterial communities in the healthy periodontium were very different from those isolated in diseased sites.
These findings contradicted the concept in vogue at the time that periodontal diseases simply resulted from an increased mass of bacteria rather than from qualitative changes in the composition of the microbiota.
At the same time, research was also exploring the nature of the inflammatory response in periodontal diseases. Numerous dynamic vascular and cellular events are activated during the onset and progression of gingival and periodontal infections, and many observations implicated PMNs as essential determinants in defending periodontal tissues against pathogenic microbes. Even in relatively healthy mouths, neutrophils migrate across the junctional epithelium into the gingival crevice where most come into direct contact with plaque microbiota, while the rest move into the oral cavity as “salivary leukocytes”.
With the development of overt infection, increased numbers of PMNs are recruited into the gingival crevice and periodontal pocket where they line up against the “plaque wall”, as if forming a barrier against microbial invasion. PMNs are the dominant inflammatory leukocytes in such a strategic position, and it was tempting to speculate that the outcome of this initial contact might be a critical determinant in disease progression. If PMNs attack and destroy microbial parasites, infection might resolve with minimal injury. On the other hand, if infection persists, irreversible injury might ensue because of sustained production and extracellular release of injurious proinflammatory factors from PMNs and from pathogenic bacteria.
Our research, at the University of Pennsylvania, investigated the effects of PMN-bacterial interactions that might be related to the pathogenesis of periodontal disease. The discovery of Y4 leukotoxicity and the subsequent identification of JP2 as a “super” leukotoxin producer were truly extraordinary events for our team – William (Bill) P. McArthur, Norton S. Taichman, Chi-Cheng Tsai, and Pierre C. Baehni from the Periodontal Research Unit, led by Max Listgarten. We came from various backgrounds, with different expertise and skills, thus forming a unique research team (see Figure 1).

Interactions of PMNs and oral microorganisms
Our early studies isolated PMNs from peripheral blood of adult donors and exposed the cells in vitro to dental plaque and specific oral gram-positive bacteria (Taichman et al. 1977). Under varying conditions, we ascertained whether these organisms interacted with PMNs, and the results showed that viable PMNs consistently phagocytosed gram-positive bacteria as well as dental plaque, as observed in vivo in the gingival crevice. Furthermore, activated human PMNs released lysosomal and other pro-inflammatory products into the culture medium and killed or inhibited growth of these plaque isolates. In the light of such findings, it seemed reasonable to postulate that our in vitro model had direct relevance to analogous events that appeared to be operating in vivo in the gingival environment.
In 1977, our research expanded to include oral gram-negative microorganisms. Sigmund Socransky at the Forsyth Dental Center in Boston identified and provided several gram-negative isolates collected from young children with periodontosis (Newman, Socransky 1977). This aggressive form of the disease attracted much attention and was frequently encountered in the periodontal clinics at the University of Pennsylvania. Back then, these organisms had not been fully characterised and were labelled according to Socransky’s laboratory as Bacteroides melaninogenicus 381, Leptotrichia buccalis 14201, Capnocytophaga 4, 6, J5, Glider 24, Fusobacterium IC 23, and Bacteroides-like Y4. Socransky later confirmed that Y4 was identified as “Actinobacillus actinomycetemcomitans”. Electron microscopic and biochemical observations showed that – with the exception of Y4 isolate – all these gram-negative organisms were consistently ingested by PMNs and triggered extracellular release of lysosomal markers into the culture medium (Tsai et al. 1978).
‘Uncharted territory’
Taking up the story, Pierre Baehni recalls: “When I examined human PMNs exposed to Y4 under the electron microscope, I observed something I had never seen before: the vast majority of PMNs appeared severely damaged, as disintegrated, and recognisable only by the general contour of the cells, as shown by the images in Figure 2 (published here for the first time). The lobulated nuclei were no longer visible and were replaced by a dense amorphous mass, while the cytoplasm presented a granular aspect and was devoid of lysosomes as well as other organelles. Only a few PMNs remained intact, but there was no evidence of phagocytic activity. I was puzzled.”
We were now in uncharted territory.
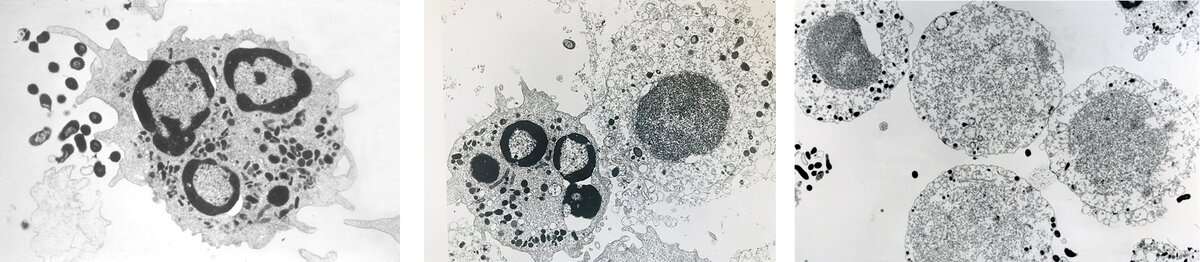
Our research group was baffled by the electron-microscopic images of shadowy, ghost-like PMNs. How could we explain what was happening? The first impulse was that something had been messed up in preparing the cell cultures or processing them for study by electron microscopy. So, the meeting concluded with “Repeat the experiment again and make it right!”
But the same observations occurred in subsequent runs: whenever they encountered Y4, PMNs appeared as mere shadows of the control cells. It was clear that these cells had been subjected to severe damage and their ghost-like appearance strongly suggested that they were no longer viable. To substantiate this hypothesis, we measured the levels of lactate dehydrogenase (LDH) released from PMNs into the culture medium. This biochemical marker of cell death was always elevated in the supernatants of the PMN/Y4 cultures but not in the controls (PMNs alone) or in PMNs incubated with gram-positive or any other gram-negative oral organisms. This was a thrilling moment. There was no longer any question or shadow of doubt: Y4 killed human PMNs.
At this exciting juncture, Bill McArthur coined the term “Y4 leukotoxin”. We reported our findings at the 1978 AADR-IADR meeting in Washington DC and the results were published the following year in Infection and Immunity (Baehni et al. 1979).
Looking back, we were lucky that theY4 isolate displayed leukotoxic activity. Several other Aa isolates or different Aa strains from the American Type Culture Collection (ATCC) failed to destroy PMNs (Baehni et al. 1981), and had we initially tested these less leukotoxic organisms, it is possible that we might never have come across Aa leukotoxin.
Antibodies neutralised Aa leukotoxin
When we immunised rabbits with crude Y4 sonic extracts, they produced high titres of serum antibodies that neutralised the leukotoxin in a dose-dependent manner. This observation prompted a series of experiments to determine whether sera from juvenile periodontitis patients also produced antibodies against the leukotoxin. And we found that whole serum as well as the immunoglobulin G fraction of patient serum neutralised leukotoxin activity – a finding that proved the existence of a direct relationship between Aa infection and the host immune response in juvenile periodontitis (McArthur et al. 1981).
In collaboration with Robert (Bob) J. Genco at the University at Buffalo, we expanded our observations and tested a panel of human sera collected from healthy donors and from patients with various oral conditions (juvenile periodontitis, adult periodontitis, acute necrotising ulcerative gingivitis, and edentulous). The results were unequivocal: almost all samples from juvenile periodontitis patients (>90%) neutralized Aa leukotoxin activity. By contrast, none of the other sera inhibited the leukotoxin (Tsai et al. 1981).
Discussing these exciting findings with Bob Genco, we hypothesised that the PMN chemotactic dysfunction reported in juvenile periodontitis combined with Aa leukotoxicity might result in the rapid development of this disease. We also speculated that vigorous production of antibodies against Aa might slow down and even halt the disease progression – a premise that could explain why the lesions in localised juvenile periodontitis were confined to the first molars and incisors.
JP2 – a new Aa strain
In the early 1980s, our group isolated a new Aa strain – referred to as JP2 – from an eight-year-old male with deep periodontal pockets and advanced alveolar bone loss affecting all his primary molars (Tsai et al. 1984). The JP2 clone belonged to the Y4 serotype and was a highly leukotoxic strain. The JP2 genotype is characterised by a specific 530-bp deletion in the promoter of the leukotoxin gene operon 28, resulting in 10- to 20-fold enhanced leukotoxic activity compared to other leukotoxic Aa strains. Crude extract and purified leukotoxin from the Aa JP2 clone shared the same properties as the Aa Y4 strain.
Aa JP2 then became the main source for leukotoxin extraction and has greatly facilitated research on the properties and the mechanisms of action of the leukotoxin. JP2 strains were found to be strongly associated with aggressive forms of periodontitis in younger individuals, particularly in North Africa and in adolescent populations of Afro-American descent. Tracing JP2 in different populations based on genetic analysis suggested that the JP2 clone carrying the mutation appeared as a distinct genotype in the Mediterranean region of Africa about 2,400 years ago and made its way to North America during the slave trade (Haubek et al. 2007).
Aa virulence attributes
Aa leukotoxin destroys human blood PMNs as well as monocytes and PMNs isolated from the gingival crevice. Aa leukotoxin also kills PMNs from non-human primates such as great apes and Old-World monkeys (Taichman et al. 1984). Interestingly, the majority of adult non-human primates harboured leukotoxic strains of Aa and produced neutralising antibodies against the toxin even though they had no signs of periodontal disease.
Our group extracted and partially purified Aa leukotoxin (Ltx) (Tsai et al. 1979, McArthur et al. 1982). Chi-Cheng Tsai demonstrated that Aa sonicate extract and partially purified Ltx bind to PMN cell membranes, while heat-treatment of Ltx (56°C, 30 min) abolished the leukotoxic effect. The DNA sequence of the leukotoxin was identified and Aa proved to produce a 114-kDa RTX (repeats-in-toxin) protein – so-called leukotoxin (LtxA). The cellular target receptor for Aa LtxA was identified as the leukocyte function antigen-1 (LFA-1) present on the cell surface. Aa LtxA disrupts the cell membrane lipid bilayer causing the death of target cells.
Since then, many other non-lethal virulence factors have been identified that may also interfere with host’s defence systems.
Why Y4?
How did this gram-negative rod come to be labelled Y4? According to Ray Williams, who was at Forsyth at that time, the plaque sample from a juvenile-periodontitis patient, was labelled with the letter Y to indicate a diseased site. The number 4 identified the colony hand-picked on the anaerobic culture plate following incubation. This so-called Y4 colony attracted Socransky’s attention because of its peculiar colony morphology.
But that is not the end of the story. We later found out that Max Listgarten had collected teeth from the very same patient who was infected with Y4. These samples were employed for his original electron microscopic studies on the structure of dental plaque (Listgarten 1976). What a twist in the story!
Aa leukotoxin – a lucky strike
Our studies and the discovery of the Y4 leukotoxin certainly sparked excitement in Actinobacillus actinomycetemcomitans as a species, paving the way to fame. Although Actinobacillus actinomycetemcomitans was later reclassified as Aggregatibacter actinomycetemcomitans, it remains “Aa” for all involved in research on this microorganism. The leukotoxin contributed to the potential importance of Aa as the prototype of a pathogen that may disrupt a critical defence system in the periodontium of infected young patients.
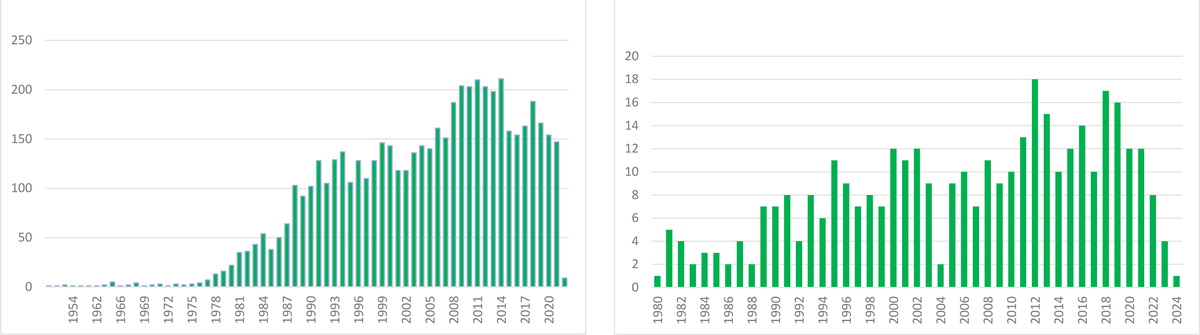
The impact of our discovery is illustrated by the number of published articles on Aa since we reported our initial findings in 1979 (search on PubMed as of 20.01.2024 According to PubMed, 5,044 papers on Aa were published between 1979 and 2024 (see Figure 3).
For our research group, this has been an exciting and rewarding adventure that will always be etched in our memories. Even after all these years, the taste of the discovery has kept its full savour!
Acknowledgments
We express our gratitude to Isao Ishikawa and William Giannobile for their encouragement in writing the story. Many thanks to Ray Williams for the amusing insights on Y4, Mogens Kilian and Panos N. Papapanou for reading the manuscript. Pierre C. Baehni is forever indebted to his mentor, Giorgio Cimasoni (1933-2008).
Bibliography
Baehni P, Tsai CC, McArthur WP, Hammond BF, Taichman NS. 1979. Interaction of inflammatory cells and oral microorganisms. VIII. Detection of leukotoxic activity of a plaque-derived Gram-negative microorganism. Infect Immun 24:233-243.
Baehni PC, Tsai CC, McArthur WP, Hammond BF, Shenker BJ, Taichman NS. 1981. Leukotoxic activity in different strains of the bacterium Actinobacillus actinomycetemcomitans isolated from juvenile periodontitis in man. Arch Oral Biol 26:671-676.
Haubek D, Poulsen K, Kilian M. Microevolution and patterns of dissemination of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect. Immun. 2007; 75:3080-3088.
Listgarten MA. 1976. Structure of the microbial flora associated with periodontal health and disease in man. J Periodontol 47:1-18.
McArthur WP, Tsai CC, Baehni PC, Genco RJ, Taichman, NS. 1981. Leukotoxic effects of Actinobacillus actinomycetemcomitans. Modulation by serum components. J. Periodontal Res 16:159-170.
McArthur WP, Tsai CC, Baehni PC, Shenker BJ, Taichman NS. 1982. Noncytolytic effects of Actinobacillus actinomycetemcomitans on leukocyte functions. In "Host-Parasite Interactions in Periodontal Diseases". p. 179-192. Eds. R.J. Genco and S.E. Mergenhagen. American Society for Microbiology. Washington, D.C
Newman MG, Socransky SS. 1977. Predominant cultivable microbiota in periodontosis. JPeriodontal Res 12:120-128.
Taichman NS, Tsai CC, Baehni PC, Stoller N, McArthur WP. 1977. Interaction of inflammatory cells and oral microorganisms. IV. In vitro release of lysosomal constituents from polymorphonuclear leukocytes exposed to supragingival and subgingival bacterial plaque. Infect Immun 16:1013-1023.
Taichman NS, Shenker BJ, Tsai CC, Glickman LT, Baehni PC, Stevens R, Hammond BF. 1984. Cytopathic effects of Actinobacillus actinomycetemcomitans on monkey blood leukocytes. J Periodontal Res 19:133-145.
Tsai CC, Hammond BF, McArthur WP, Baehni P, Taichman NS. 1978. Interaction of inflammatory cells and oral microorganisms. VI. Exocytosis of PMN lysosomal products from human and rabbit polymorphonuclear leukocytes exposed to Gram-negative plaque pathogens. J Periodontal Res 13:504-512.
Tsai CC, McArthur WP, Baehni PC, Hammond BF, Taichman NS. 1979. Extraction and partial characterization of a leukotoxin from a plaque-derived microorganism. Infect Immun. 24:427-439.
Tsai CC, McArthur, WP, Baehni, PC, Evian C, Genco, RJ, Taichman NS. 1981. Serum neutralizing activity against Actinobacillus actinomycetemcomitans leukotoxin in juvenile periodontitis. J Clin Periodontol 8:338-348.
Tsai CC, Shenker BJ, DiRienzo JM, Malamud D, Taichman NS. 1984. Extraction and isolation of a leukotoxin from Actinobacillus actinomycetemcomitans with polymyxin B. Infect Immun 43:700-705.
Biographies
Pierre C. Baehni is professor emeritus of the Faculty of Medicine, University of Geneva, Switzerland. After obtaining a doctorate from the University of Geneva, in 1974 he joined the University of Pennsylvania, Department of Periodontics, School of Dental Medicine, as research fellow and later as assistant professor. From 1983-2011, he was professor at the School of Dental Medicine, University of Geneva. He was a senior consultant at the WHO (2010-2015) and served as president of the Swiss Society of Periodontology (1989-1992), president of the EFP (1994-1995), and secretary general of the EFP (2006-2010).
William P. McArthur is professor emeritus of the University of Florida, USA. After completing his PhD at Purdue University, he was a post-doctoral fellow at the New York University Medical School and then became associate professor at the University of Pennsylvania School of Dental Medicine (1972-1981). In 1981, he joined the University of Florida as professor and chairman of the Department of Oral Biology. Later, he became assistant dean for research followed by associate dean for faculty affairs, at the University of Florida College of Dentistry.
Norton S. Taichman earned a DDS from the Faculty of Dentistry, University of Toronto, Canada in 1961. He then spent three years in Boston at the Harvard School of Dental Medicine focusing on periodontics and pathology. After completing his PhD in pathology at the University of Toronto, Faculty of Medicine, he was promoted to associate professor of pathology in Toronto’s Dental and Medical schools. From 1972 to 2001, he was professor and chairman of pathology at the University of Pennsylvania, School of Dental Medicine in Philadelphia.
Chi-Cheng Tsai is Professor Emeritus at the Chun Shan Medical University. He completed his PhD in immunopathology at Toronto University, Canada, and was research associate professor at the University of Pennsylvania, School of Dental Medicine, USA. He was the first dean of the College of Dental Medicine, Kaohsiung Medical University, Taiwan. He has been president of the Taiwan Association for Dental Sciences, of the Taiwan Academy of Periodontology, and is the recipient of an award for his work in periodontal-dental sciences.